News
Preferred Networks releases ChainerX, a C++ implementation of automatic differentiation of N-dimensional arrays, integrated into Chainer v6 (beta version) for higher computing performance
2018.12.03
Dec. 3, 2018, Tokyo Japan – Preferred Networks, Inc. (“PFN”, Head Office: Tokyo, President & CEO: Toru Nishikawa) releases ChainerX, a C++ implementation of automatic differentiation of N-dimensional arrays for the Chainer™ v6 open source deep learning framework. Chainer v6 will run without the need to change most of the code used in previous versions.
Since the release of its source code in 2015, the development of Chainer, known as a pioneer of flexible and intuitive deep learning frameworks, has been very active and attracted many users. Many other deep learning frameworks have followed suit in adopting Chainer’s Define-by-Run method, demonstrating the foresight of Chainer. Chainer’s pure Python implementation policy has, on the one hand, contributed to the legibility and simplicity of code, but on the other, it was becoming a bottleneck due to increased overhead of the Python execution system relative to the overall runtime as its performance improved.
Therefore, the release of ChainerX, which is written in C++ and integrated into the main Chainer, is a first step in achieving higher performance without losing much of Chainer’s flexibility and backward compatibility for many users.

Main features of ChainerX are:
- C++ implementation in close connection with Python – NumPy, CuPy™, and automatic differentiation (autograd), all of which are mostly written in Python, have been implemented in C++
The logic of matrix calculation, convolution operations, and error backpropagation has all been implemented in C++ to reduce CPU overhead by Python by up to 87% (comparison of overhead measurements only)
- Easy to work with CPU, GPU, and other hardware backends
Replaceable backends have increased portability between devices
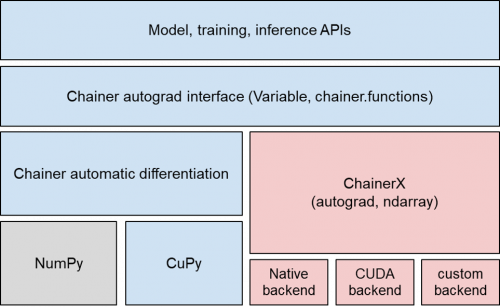
Figure:In addition to the multidimensional array implementation which corresponds to NumPy/CuPy, the Define-by-Run style automatic differentiation function is covered by ChainerX.
- Chainer Release Note: https://github.com/chainer/chainer/releases/tag/v6.0.0b1
- ChainerX Documentation: https://docs.chainer.org/en/latest/chainerx/index.html
- Blog: https://chainer.org/announcement/2018/12/03/chainerx.html
As well as improving ChainerX performance and expanding the backend, PFN plans to enable models written in ChainerX to be called from non-Python environments.
For more details on Chainer X, developer Seiya Tokui is scheduled to give a presentation at NeurIPS, a top conference in machine learning (formerly called NIPS), in Montreal, Canada this month.
Dec. 7, 12:50-02:55 Open Source Software Showcase:
http://learningsys.org/nips18/schedule.html
Chainer has adopted a number of development proposals from external contributors. PFN will continue to quickly adopt the results of the latest deep learning research and promote the development and popularization of Chainer in collaboration with supporting companies and the OSS community.
- About the Chainer™ Open Source Deep Learning Framework
Chainer is a Python-based deep learning framework developed and provided by PFN, which has unique features and powerful performance that allow for designing complex neural networks easily and intuitively, thanks to its “Define-by-Run” approach. Since it was open-sourced in June 2015, as one of the most popular frameworks, Chainer has attracted not only the academic community but also many industrial users who need a flexible framework to harness the power of deep learning in their research and real-world applications.
Chainer quickly incorporates the results of the latest deep learning research. With additional packages such as ChainerRL (reinforcement learning), ChainerCV (computer vision), and Chainer Chemistry(a deep learning library for chemistry and biology)and through the support of Chainer development partner companies, PFN aims to promote the most advanced research and development activities of researchers and practitioners in each field. (http://chainer.org/)


